With the rapid growth of high-throughput RNA-seq and other omics data, there is an increasing need for tools to analyse the features of large lncRNA sets, such as for example those differentially expressed between two conditions.
LnCompare is a web server for lncRNA feature analysis. It is based on the GENCODE v24 lncRNA annotation, comprising 15,941 loci, for which >20 useful features have been compiled. It comprises two functonal module.
1) Gene set feature comparison. It compares two sets of lncRNAs, and identifies significantly different features, both categorical and quantitative.
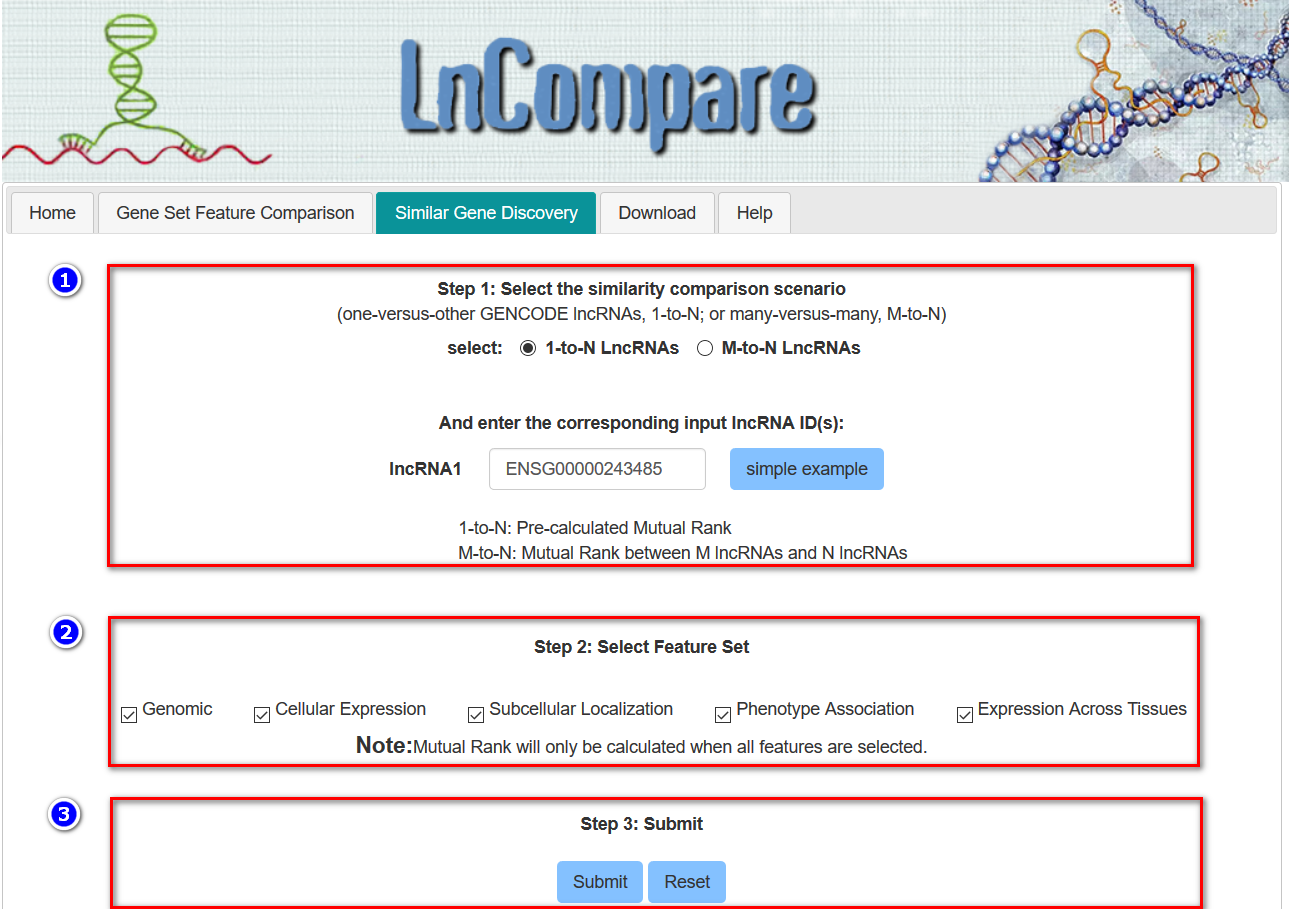
2) Similar gene discovery. It uses user-defined gene(s)-of-interest to query for the set of most similar lncRNAs, based on one or more features.
All analyses are performed at the levels of gene loci (not transcripts). The full feature datasets can also be downloaded in the "Download" page.
Comprehensive usage instructions can be found in the "Help" menu.
Jul 2018: the original LnCompare program is released.
Feb 2019: a stable similar gene discovery module has been established.
Dr. Rory Johnson, Department of Biomedical Research, University of Bern 3008 Bern, Switzerland.
Email: roryjohnson.dkf@gmail.com
Dr. Jianwei Li, school of artificial intelligence, Hebei University of Technology, Tianjin 300401, China
Email: lijianwei@hebut.edu.cn
Dr. Yuan Zhou, Department of Biomedical Informatics, School of Basic Medical Sciences, Peking University, Beijing, China
Email: soontide6825@163.com
LnCompare works on GENCODE lncRNA gene loci (ENSGxxxxxxxxxxx), not lncRNA transcripts.
This website has been tested by using Chrome, Microsoft Edge and Firefox browsers. Microsoft IE may not work well.
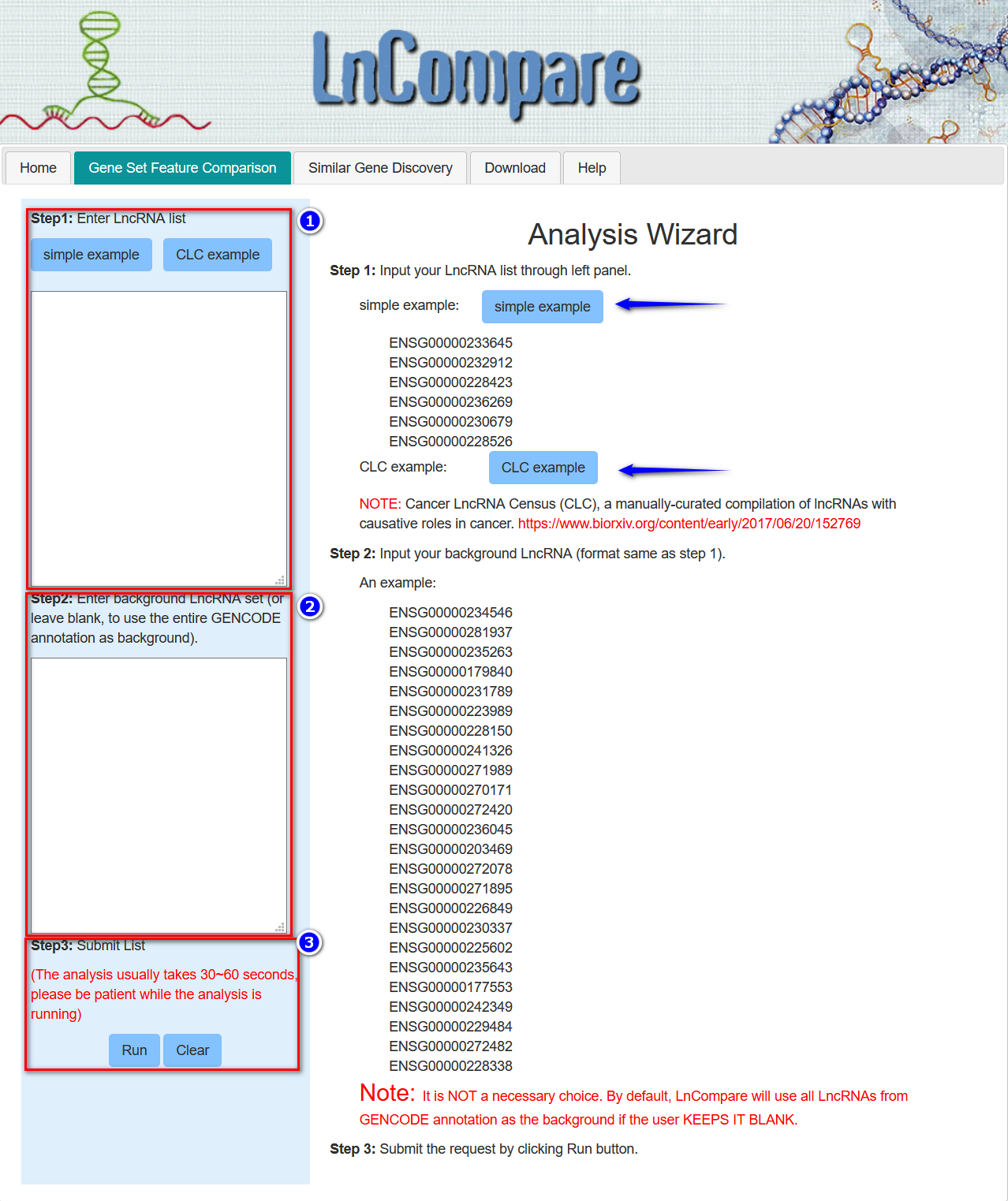
Step 1: Input your LncRNA list through left panel.
simple example:
CLC example:
NOTE: Cancer LncRNA Census (CLC), a manually-curated compilation of lncRNAs with causative roles in cancer. https://www.biorxiv.org/content/early/2017/06/20/152769
cell cycle example:
NOTE:It is a positive control dataset of 117 lncRNAs differentially expressed between G1S and G2M cell cycle phases in human cells.
Step 2: Input your background LncRNA (format same as step 1).
An example:
Note: It is NOT a necessary choice. By default, LnCompare will use all LncRNAs from GENCODE annotation as the background if the user KEEPS IT BLANK. Also, please ensure that the background list has more lncRNAs than the input lncRNA list.
Step 3: Submit the request by clicking Run button.
Download the entire feature table for LnCompare here: table_feature.
How to use and interpret the Similar Gene Discovery function?
Note that, since all features are determined for GENCODE annotated lncRNA genes, every lncRNA should be present in GENCODE and should be passed using the corresponding GENCODE identifier, which begins with “ENSG”. The user may also start two kinds of pre-defined lncRNA examples by clickcing the corresponding buttons: a simple lncRNA or Cancer LncRNA Census (CLC), a manually-curated compilation of LncRNAs with causative roles in cancer. https://www.biorxiv.org/content/early/2017/06/20/152769
Note: If no background list is provided, then by default, LnCompare will use the entire GENCODE annotation set as the background.
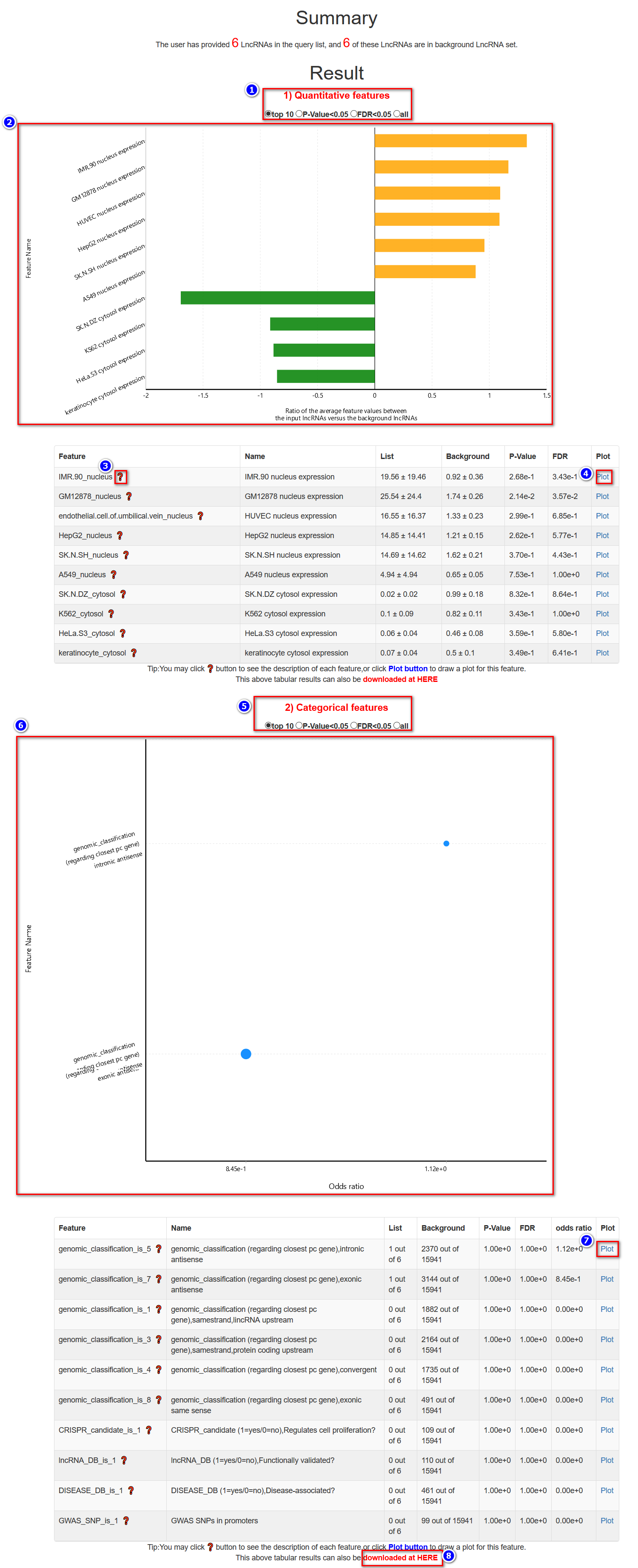
A brief summary of the user's input is provided (indicated by the green lines below).
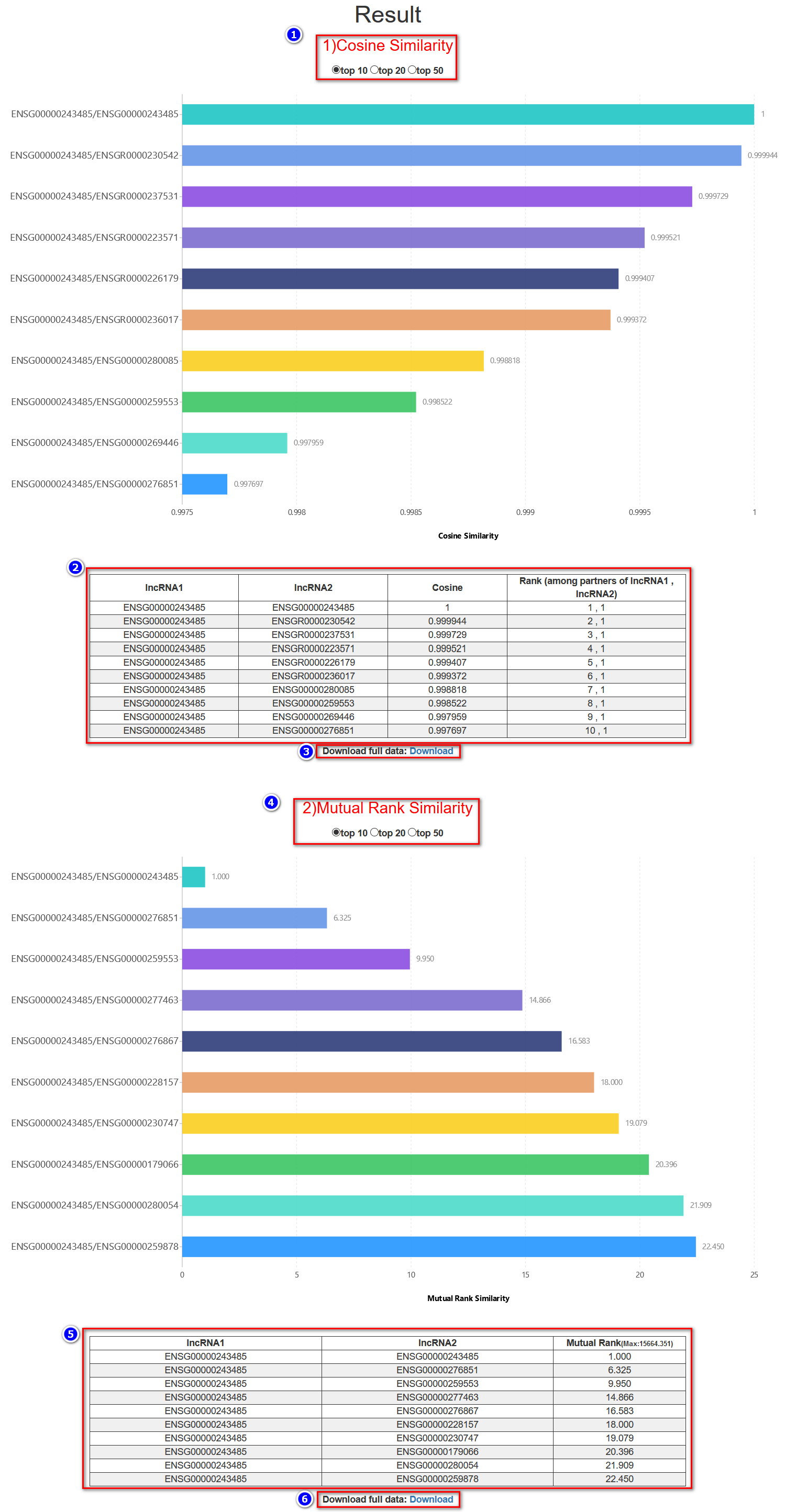
In this tabular result page, users are provided with two kinds of analysis results: quantitative features and categorical features.
(---For quantitative features---)
(---For categorical features---)
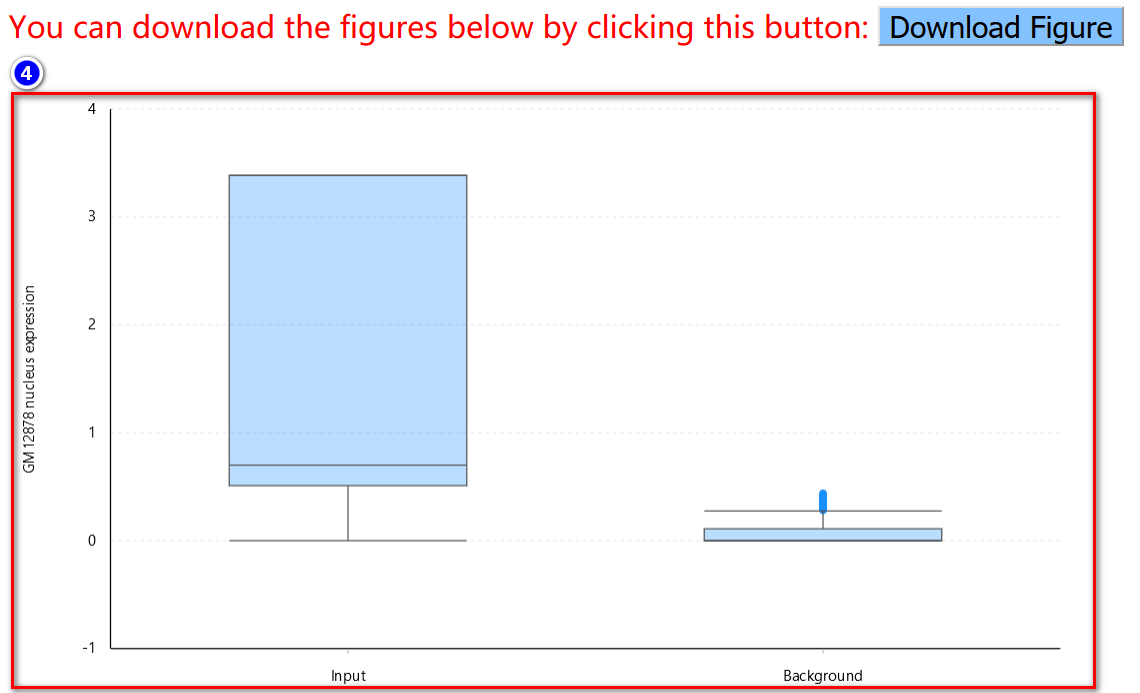
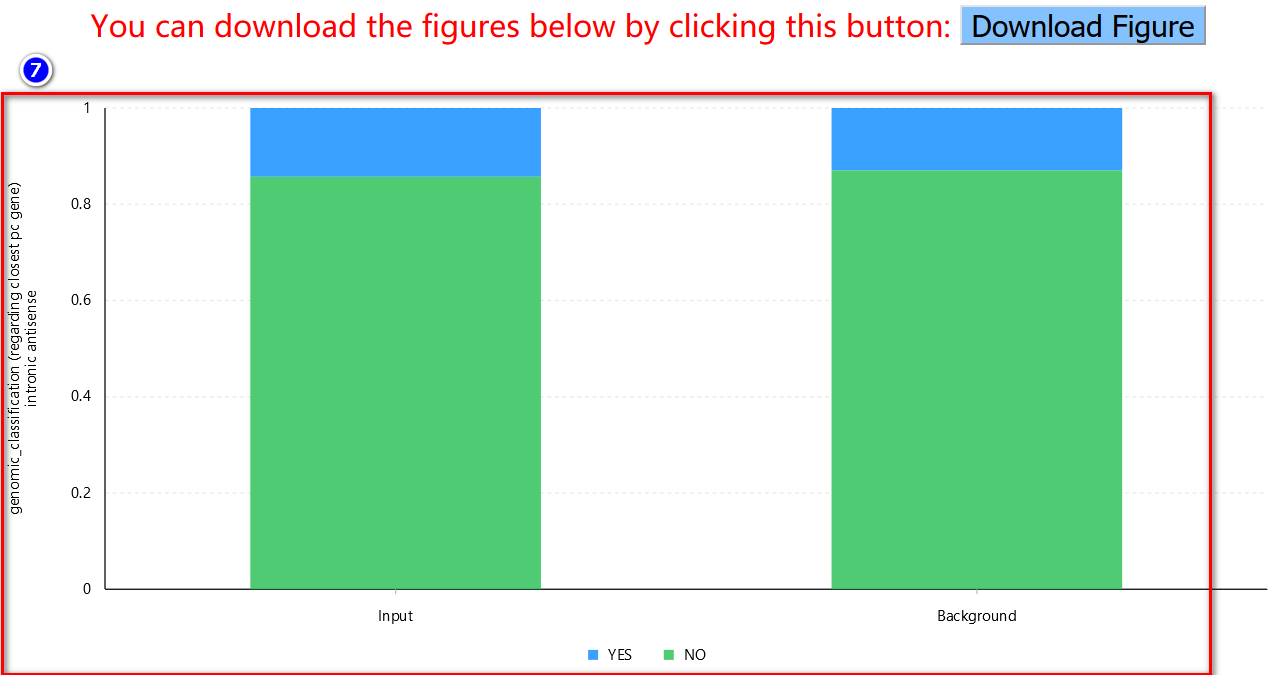
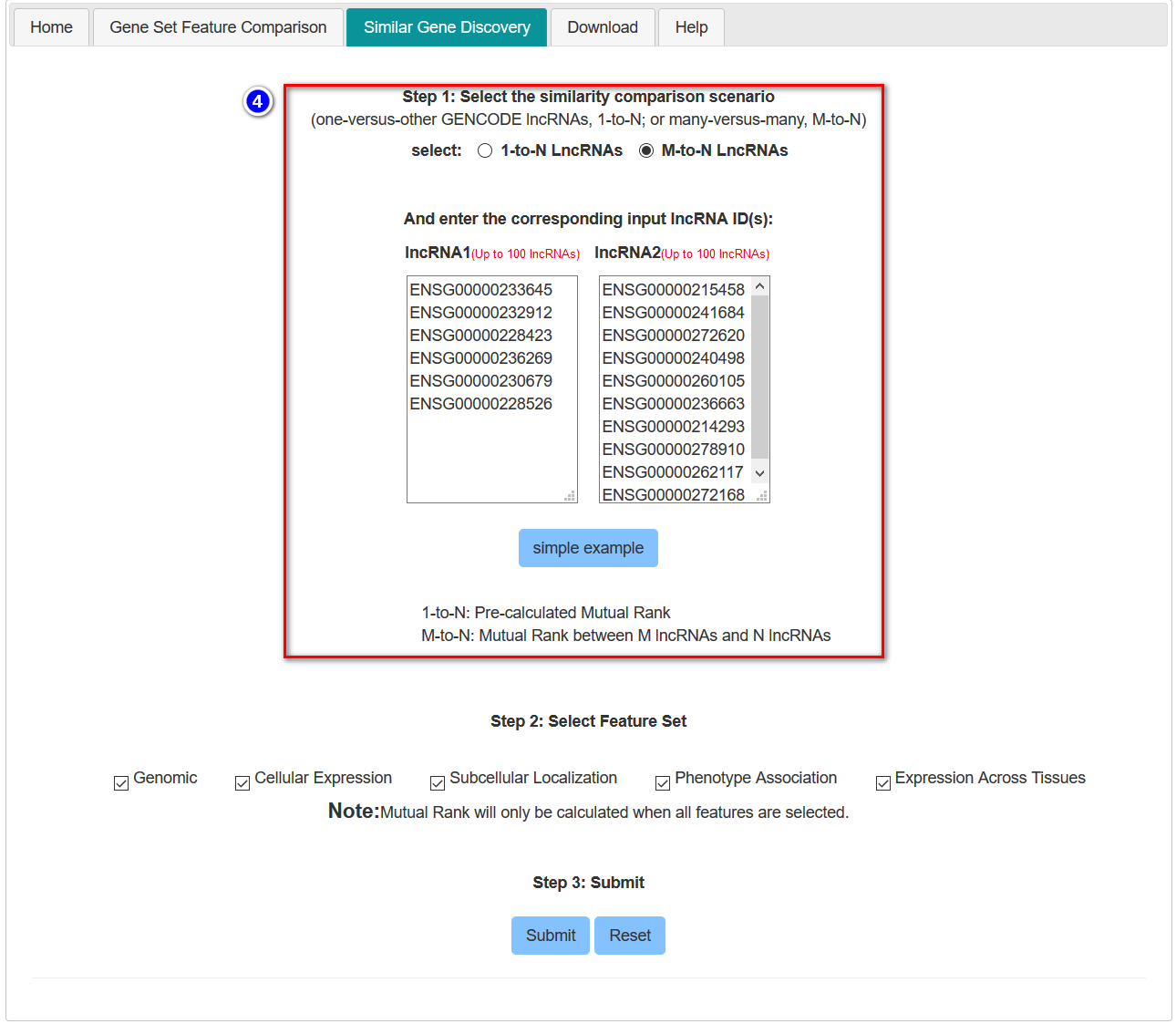
Note: If users choose the M-to-N comparison, the maximum size of each set is 100. Due to intensive computational requirements for the mutual rank similarity, when performing 1-to-N comparison, the pre-calculated mutual rank similarity will be retrieved only if all five feature subsets are enabled (default option).
The entire feature table of LnCompare can be found on the download page, user can click 'table_feature' link to download it.